Omics-Based Dissection of Alternative Splicing and the Regulatory Architecture in Fungal Genomes
- David Ojcius
- Jan 20
- 2 min read
Highlights
Alternative splicing has been implicated in the regulation of a variety of genes
Intron retention is the most frequent alternative splicing event in fungi
We critically identified the gaps in the role of alternative splicing in fungi
We proposed a practical roadmap to fill these gaps.
ABSTRACT
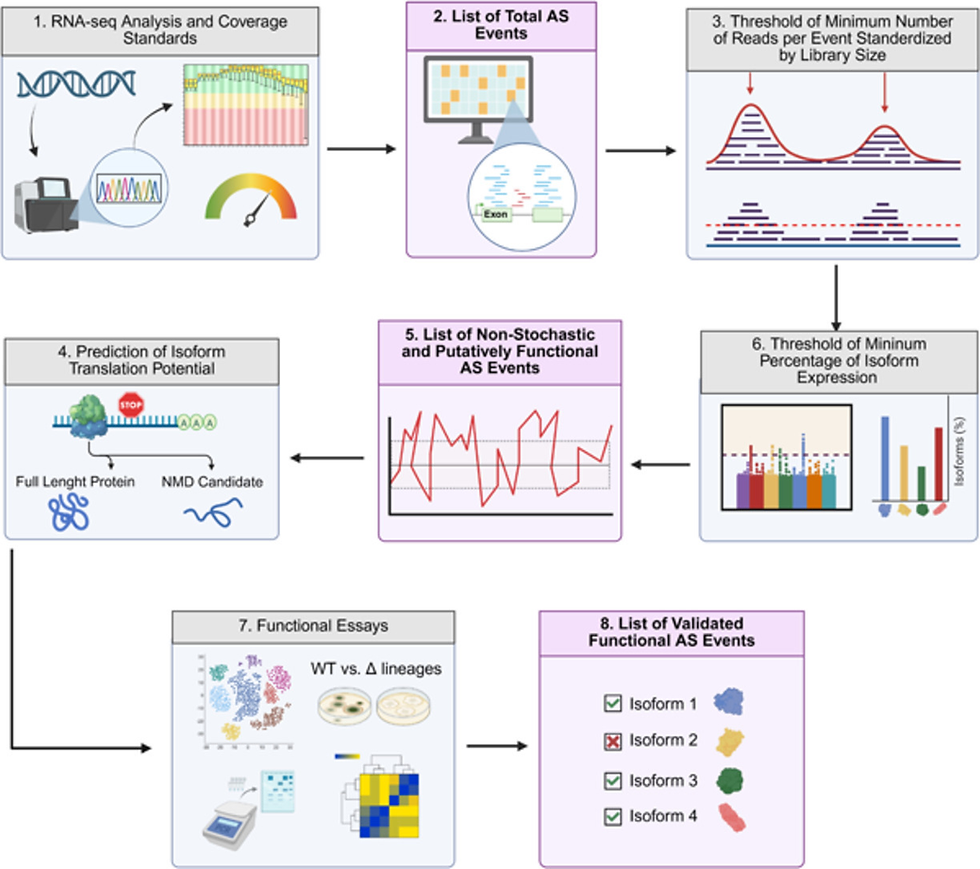
Assigning mechanistic and functional associations to most of the widespread splicing diversity in fungi has been one of the continuous and most intriguing challenges in the field of molecular mycology. Ongoing advances in sequencing technologies have driven recent outbreaks in the study of the dynamic splicing landscapes of fungal transcriptomes. Nevertheless, most ascomycete and basidiomycete species of agronomic, industrial, and medical interest exhibit a notable proportion of their transcripts affected by alternative splicing (AS), producing mRNA isoforms whose regulation and functions have been largely ignored to date. In contrast to all other fungi, some yeast and basal fungal clades exhibit extremely scarce AS occurrences, which are associated with very low genomic intron counts. In this review, we provide thoughtful insights into three long-standing unsolved questions: (1) the pervasive occurrence and possible stochasticity of intron retention in fungi, (2) the reasons for increased AS rates in pathogenic and filamentous species, and (3) possible hypotheses to explain the functionality and fitness advantages of AS in fungi. We present a critical discussion on the role of AS, highlighting its importance in an evolutionary context that integrates major features of fungal diversification and adaptability. To advance our understanding of fungal splicing control, function, and evolution, we propose a practical roadmap for future research, aimed at filling the most critical gaps in the field.
Read article for free (open access):




Comments